Dr Rebecca (Beky) Weiser is a Postdoctoral Research Associate at Cardiff University, she is currently investigating the diversity and evolution of the lung microbiota in children with Cystic Fibrosis (CF). Beky works closely with Professor Esh Mahenthiralingam, who sits on the CF AMR Syndicate Steering Committee, and Dr Julian Forton. In this blog post, we caught up with Beky to find out more about her research.
Beky, can you tell us a little about your research; what are the challenges you’re trying to address?
I am currently working as a researcher-co-investigator on a project funded by the US Cystic Fibrosis Foundation to investigate the lung microbiota in children with CF. This is challenging as children with CF often do not expectorate sputum even when unwell, meaning that other methods of sampling are needed to detect if they have microbial infections. Current routinely used sampling methods for children have limitations, as cough swabs are easy to perform but insensitive, and the gold standard sampling technique bronchoalveolar lavage (BAL) is invasive (it requires general anaesthetic) so cannot be performed frequently.
Working with the Cystic Fibrosis Sputum Induction Trial (CF-SpIT), we are evaluating the performance of induced sputum (IS) sampling for detecting specific antimicrobial resistant pathogens and the microbiota in the lower airways. As IS is non-invasive and can performed frequently, it can be used to collect multiple longitudinal samples. If IS can accurately capture the lung microbiota, we can use it to aid pathogen diagnosis, build up a picture of microbiota evolution and then start to understand the short- and long- term effects of antibiotic treatment on lung infections. This is really exciting as current sampling techniques for children with CF cannot be used in this way.
What results have you generated so far?
The previous culture microbiology work of CF-SpIT trial revealed that IS outperformed cough swab and matched the performance of the gold standard BAL in terms of pathogen detection. My project extends this work by using sensitive culture-independent (DNA based) techniques such as microbiota analysis to identify the different bacterial species in matched IS and BAL samples. So far, by looking at BAL samples we have found that microbiota compartmentalisation can occur in the lower airways of children with CF. This means that different bacterial communities are found in different lobes of the lung and presents problems for consistent microbiological sampling. We have also found that in approximately 80% of cases, IS captures a microbiota signature representative of one or more of the BAL samples taken from the lower airways, identifying the dominant or prevalent bacterial species. These results are promising, indicating that IS can reliably aid pathogen diagnosis and be used to understand microbiota evolution, even when lower airway samples may be variable.
How does your research relate to or impact on people with CF?
The CF-SpIT study has already had an impact on CF care, with many centres in the UK now performing IS routinely and other centres in Europe, the USA and Australia reconsidering their approaches to regular sampling in children. Using IS sampling and molecular diagnostics we will be able to improve pathogen detection and understand the impact of different treatments (including antibiotics) on the microbiota and disease progression. In addition, as many adults with CF who are taking CFTR modulator therapies are now expectorating less, IS may present a viable alternative to sputum samples to diagnose and monitor lung infections in adults.
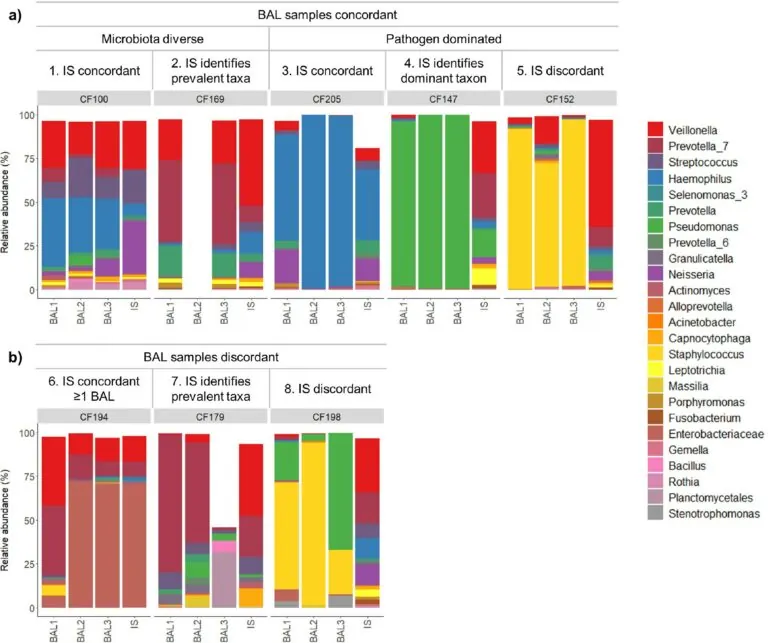
Fig 1: The different microbiota profile types found by our study. This figure shows the 8 different microbiota profile types found from the 30 sample sets in the study. Samples are grouped by sample set number and whether BAL samples are (a) concordant (similar), or (b) discordant (dissimilar). The performance of IS in relation to BAL samples is also noted. The top 25 most abundant genera/families across the sample set are indicated in the colour key. In 24/30 (80%) of cases, IS captured a microbiota signature representative of one or more of the BAL samples taken from the lower airways, identifying the dominant or prevalent bacterial species (profile types 1, 2, 3, 4, 6 and 7).
Full findings from Beky’s research have been published in the Journal of Cystic Fibrosis and can be accessed here.
What’s next for your research?
As you may have expected from the above results, we are now analysing a large collection of IS samples, including longitudinal samples, to investigate microbiota evolution and correlate bacterial diversity to clinical parameters such as antibiotic treatments. At the same time, we are also using microbiota analysis to look at the fungal communities in CF-SpIT samples. This is really interesting and important as relatively little is known about the prevalence or clinical impact of fungi in CF, particularly in children.
How did you get into CF antimicrobial research, and why do you find it so interesting?
All of my research projects have had some link to CF research, whether it was researching antibiotic resistant bacterial species that are common pathogens in CF lung infections (Pseudomonas aeruginosa and Burkholderia cepacia complex species) or assisting clinical trials by performing microbiota analysis on CF samples. I have also worked with a company, AlgiPharma AS, who were carrying out a clinical trial on a new therapeutic, Oligo G, for these antibiotic resistant bacteria. I enjoy working with different collaborators and the translational aspect of the work I have done. It is great to think that something you are researching can positively impact the lives of people with CF.
I also find the data analysis process really interesting! I only really started learning bioinformatics and statistics for microbiota analysis during my first postdoctoral position. This was quite daunting as I hadn’t previously thought of myself as being particularly good at these things! Learning these skills has really opened my eyes to the world of data analysis, particularly to the tools and approaches used for analysing large datasets such as those used in microbiota research. I really enjoy exploring the data and working out the best approach to answer my research questions.
How important is collaboration in your work? Are there any areas of research/capabilities in particular that you’re looking to collaborate in?
As this research is very translational, collaboration has been extremely important at every stage, from sample collection and processing to data analysis and results interpretation. We have regular meetings as a project team to ensure that we are asking the right questions in our analysis and that we can understand the clinical significance of the results. It has been a really useful experience to see how academic research can inform clinical practice.
As microbiota analysis is a complex and dynamic area of research, I hope that in the future I can collaborate with other bioinformaticians and statisticians to ensure that I am using the most up-to-date tools and models in my work. In particular, I would like to use microbiota analysis to look at co-occurrence of different species in CF lung infections and generate hypotheses about species interactions that can be tested in vitro in the lab.